- Record: found
- Abstract: found
- Article: not found
Competitive regulation of synaptic Ca influx by D2 dopamine and A2A adenosine receptors
Abstract
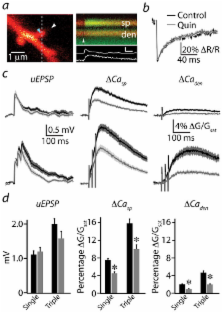
Striatal D2-type dopamine receptors (D2Rs) are implicated in the pathophysiology of neuropsychiatric disorders, including Parkinson’s disease and schizophrenia. Although these receptors regulate striatal synaptic plasticity, the mechanisms underlying dopaminergic modulation of glutamatergic synapses are unclear. We combined optogenetics, 2-photon microscopy, and glutamate uncaging to examine D2R-dependent modulation of glutamatergic synaptic transmission in mouse striatopallidal neurons. We find that D2R activation reduces corticostriatal glutamate release and attenuates both synaptic- and action potential-evoked Ca influx into dendritic spines by approximately 50%. Modulation of Ca signaling is mediated by a PKA-dependent regulation of Ca entry through NMDA-type glutamate receptors that is inhibited by D2Rs and enhanced by activation of 2A-type adenosine receptors (A2ARs). D2Rs also produce a PKA- and A2AR-independent reduction in Ca influx through R-type voltage-gated Ca channels. These findings reveal that dopamine regulates spine Ca by multiple pathways and that competitive modulation of PKA controls NMDAR-mediated Ca signaling in the striatum.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons.
- Record: found
- Abstract: found
- Article: not found
Dopamine receptors: from structure to function.
- Record: found
- Abstract: found
- Article: not found
