- Record: found
- Abstract: found
- Article: found
An Epstein-Barr virus–encoded microRNA targets PUMA to promote host cell survival
Read this article at
Abstract
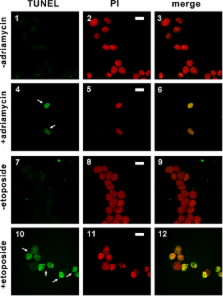
Epstein-Barr virus (EBV) is a herpesvirus associated with nasopharyngeal carcinoma (NPC), gastric carcinoma (GC), and other malignancies. EBV is the first human virus found to express microRNAs (miRNAs), the functions of which remain largely unknown. We report on the regulation of a cellular protein named p53 up-regulated modulator of apoptosis (PUMA) by an EBV miRNA known as miR-BART5, which is abundantly expressed in NPC and EBV-GC cells. Modulation of PUMA expression by miR-BART5 and anti–miR-BART5 oligonucleotide was demonstrated in EBV-positive cells. In addition, PUMA was found to be significantly underexpressed in ∼60% of human NPC tissues. Although expression of miR-BART5 rendered NPC and EBV-GC cells less sensitive to proapoptotic agents, apoptosis can be triggered by depleting miR-BART5 or inducing the expression of PUMA. Collectively, our findings suggest that EBV encodes an miRNA to facilitate the establishment of latent infection by promoting host cell survival.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
PUMA induces the rapid apoptosis of colorectal cancer cells.
- Record: found
- Abstract: found
- Article: not found