- Record: found
- Abstract: found
- Article: not found
Adaptive boosting-based computational model for predicting potential miRNA-disease associations
Abstract
Motivation
Recent studies have shown that microRNAs (miRNAs) play a critical part in several biological processes and dysregulation of miRNAs is related with numerous complex human diseases. Thus, in-depth research of miRNAs and their association with human diseases can help us to solve many problems.
Results
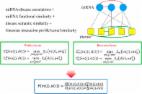
Due to the high cost of traditional experimental methods, revealing disease-related miRNAs through computational models is a more economical and efficient way. Considering the disadvantages of previous models, in this paper, we developed adaptive boosting for miRNA-disease association prediction (ABMDA) to predict potential associations between diseases and miRNAs. We balanced the positive and negative samples by performing random sampling based on k-means clustering on negative samples, whose process was quick and easy, and our model had higher efficiency and scalability for large datasets than previous methods. As a boosting technology, ABMDA was able to improve the accuracy of given learning algorithm by integrating weak classifiers that could score samples to form a strong classifier based on corresponding weights. Here, we used decision tree as our weak classifier. As a result, the area under the curve (AUC) of global and local leave-one-out cross validation reached 0.9170 and 0.8220, respectively. What is more, the mean and the standard deviation of AUCs achieved 0.9023 and 0.0016, respectively in 5-fold cross validation. Besides, in the case studies of three important human cancers, 49, 50 and 50 out of the top 50 predicted miRNAs for colon neoplasms, hepatocellular carcinoma and breast neoplasms were confirmed by the databases and experimental literatures.
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
Circulating Exosomal microRNAs as Biomarkers of Colon Cancer
- Record: found
- Abstract: found
- Article: not found
RWRMDA: predicting novel human microRNA-disease associations.
- Record: found
- Abstract: found
- Article: found