- Record: found
- Abstract: not found
- Article: not found
Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5
letter
Nicole P. Hachmann , B.S.,
Jessica Miller , B.S.,
Ai-ris Y. Collier , M.D.,
John D. Ventura , Ph.D.,
Jingyou Yu , Ph.D.,
Marjorie Rowe , B.S.,
Esther A. Bondzie , M.S.N.,
Olivia Powers , B.S.,
Nehalee Surve , M.S.,
Kevin Hall , B.S.,
Dan H. Barouch , M.D., Ph.D.
22 June 2022
Keyword part (code): 18Keyword part (keyword): Infectious DiseaseKeyword part (code): 18_2Keyword part (keyword): VaccinesKeyword part (code): 18_6Keyword part (keyword): Viral InfectionsKeyword part (code): 18_12Keyword part (keyword): Coronavirus ,
18,
Infectious Disease,
Keyword part (code): 18_2Keyword part (keyword): VaccinesKeyword part (code): 18_6Keyword part (keyword): Viral InfectionsKeyword part (code): 18_12Keyword part (keyword): Coronavirus ,
18_2,
Vaccines,
18_6,
Viral Infections,
18_12,
Coronavirus
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
To the Editor: In recent months, multiple lineages of the omicron (B.1.1.529) variant
of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have emerged,
1
with subvariants BA.1 and BA.2 showing substantial escape from neutralizing antibodies.
2-5
Subvariant BA.2.12.1 is now the dominant strain in the United States, and BA.4 and
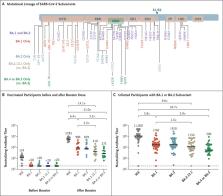
BA.5 are dominant in South Africa (Figure 1A). Subvariants BA.4 and BA.5 have identical
sequences of the spike protein.
We evaluated neutralizing antibody titers against the reference WA1/2020 isolate of
SARS-CoV-2 along with omicron subvariants BA.1, BA.2, BA.2.12.1, and BA.4 or BA.5
in 27 participants who had been vaccinated and boosted with messenger RNA vaccine
BNT162b2 (Pfizer–BioNTech) and in 27 participants who had been infected with the BA.1
or BA.2 subvariant a median of 29 days earlier (range, 2 to 113) (Tables S1 and S2
in the Supplementary Appendix, available with the full text of this letter at NEJM.org).
In the vaccine cohort, participants were excluded if they had a history of SARS-CoV-2
infection or a positive result on nucleocapsid serologic analysis or if they had received
another vaccine against coronavirus disease 2019 (Covid-19) or an immunosuppressive
medication.
Six months after the initial two BNT162b2 immunizations, the median neutralizing antibody
pseudovirus titer was 124 against WA1/2020 but less than 20 against all the tested
omicron subvariants (Figure 1B). Two weeks after administration of the booster dose,
the median neutralizing antibody titer increased substantially, to 5783 against the
WA1/2020 isolate, 900 against the BA.1 subvariant, 829 against the BA.2 subvariant,
410 against the BA.2.12.1 subvariant, and 275 against the BA.4 or BA.5 subvariant.
These data show that as compared with the response against the WA1/2020 isolate, the
neutralizing antibody titer was lower by a factor of 6.4 against BA.1, by a factor
of 7.0 against BA.2, by a factor of 14.1 against BA.2.12.1, and by a factor of 21.0
against BA.4 or BA.5. In addition, as compared with the median neutralizing antibody
titer against the BA.1 subvariant, the median titer was lower by a factor of 2.2 against
the BA.2.12.1 subvariant and by a factor of 3.3 against the BA.4 or BA.5 subvariant.
Among the participants who had been infected with the BA.1 or BA.2 subvariant of omicron,
all but one had been vaccinated against Covid-19. Because of the variation in sampling
after the onset of infection, some samples may not reflect peak neutralizing antibody
titers (Table S2). Among the participants with a history of Covid-19, the median neutralizing
antibody titer was 11,050 against the WA1/2020 isolate, 1740 against the BA.1 subvariant,
1910 against the BA.2 subvariant, 1150 against the BA.2.12.1 subvariant, and 590 against
the BA.4 or BA.5 subvariant (Figure 1C). These data show that as compared with the
WA1/2020 isolate, the median neutralizing antibody titer was lower by a factor of
6.4 against BA.1, by a factor of 5.8 against BA.2, by a factor of 9.6 against BA.2.12.1,
and by a factor of 18.7 against BA.4 or BA.5. In addition, as compared with the median
titers against the BA.1 subvariant, the median titer was lower by a factor of 1.5
against the BA.2.12.1 subvariant and by a factor of 2.9 against the BA.4 or BA.5 subvariant.
These data show that the BA.2.12.1, BA.4, and BA.5 subvariants substantially escape
neutralizing antibodies induced by both vaccination and infection. Moreover, neutralizing
antibody titers against the BA.4 or BA.5 subvariant and (to a lesser extent) against
the BA.2.12.1 subvariant were lower than titers against the BA.1 and BA.2 subvariants,
which suggests that the SARS-CoV-2 omicron variant has continued to evolve with increasing
neutralization escape. These findings provide immunologic context for the current
surges caused by the BA.2.12.1, BA.4, and BA.5 subvariants in populations with high
frequencies of vaccination and BA.1 or BA.2 infection.
Related collections
Most cited references5

- Record: found
- Abstract: found
- Article: found
Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa
Raquel Viana, Sikhulile Moyo, Daniel G. Amoako … (2022)

- Record: found
- Abstract: found
- Article: found
Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization
Sandile Cele, Laurelle Jackson, David Khoury … (2021)
- Record: found
- Abstract: found
- Article: not found
Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2
Lihong Liu, Sho Iketani, Yicheng Guo … (2021)
