- Record: found
- Abstract: found
- Article: found
Challenges Predicting Ligand-Receptor Interactions of Promiscuous Proteins: The Nuclear Receptor PXR
Read this article at
Abstract
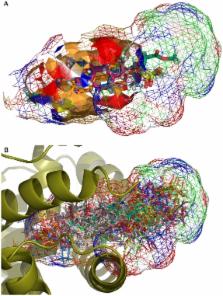
Transcriptional regulation of some genes involved in xenobiotic detoxification and apoptosis is performed via the human pregnane X receptor (PXR) which in turn is activated by structurally diverse agonists including steroid hormones. Activation of PXR has the potential to initiate adverse effects, altering drug pharmacokinetics or perturbing physiological processes. Reliable computational prediction of PXR agonists would be valuable for pharmaceutical and toxicological research. There has been limited success with structure-based modeling approaches to predict human PXR activators. Slightly better success has been achieved with ligand-based modeling methods including quantitative structure-activity relationship (QSAR) analysis, pharmacophore modeling and machine learning. In this study, we present a comprehensive analysis focused on prediction of 115 steroids for ligand binding activity towards human PXR. Six crystal structures were used as templates for docking and ligand-based modeling approaches (two-, three-, four- and five-dimensional analyses). The best success at external prediction was achieved with 5D-QSAR. Bayesian models with FCFP_6 descriptors were validated after leaving a large percentage of the dataset out and using an external test set. Docking of ligands to the PXR structure co-crystallized with hyperforin had the best statistics for this method. Sulfated steroids (which are activators) were consistently predicted as non-activators while, poorly predicted steroids were docked in a reverse mode compared to 5α-androstan-3β-ol. Modeling of human PXR represents a complex challenge by virtue of the large, flexible ligand-binding cavity. This study emphasizes this aspect, illustrating modest success using the largest quantitative data set to date and multiple modeling approaches.
Author Summary
Promiscuous proteins generally bind a large array of diverse ligand structures. This may be facilitated by a very large binding site, multiple binding sites, or a flexible binding site that can adjust to the size of the ligand. These aspects also increase the complexity of predicting whether a molecule will bind or not to such proteins which frequently function as exogenous compound sensors to respond to toxic stress. For example, transporters may prevent absorption of some molecules, and enzymes may convert them to more readily excretable compounds (or alternatively activate them prior to further clearance by other detoxification enzymes). Nuclear hormone receptors may respond to ligands and then affect downstream gene expression to upregulate both enzymes and transporters to increase the clearance for the same or different molecules. We have assessed the ability of many different ligand-based and structure-based computational approaches to model and predict the activation of human PXR by steroidal compounds. We find the most effective computational approach to identify potential steroidal PXR agonists which are clinically relevant due to their widespread use in clinical medicine and the presence of mimics in the environment.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway.
- Record: found
- Abstract: found
- Article: not found
SXR, a novel steroid and xenobiotic-sensing nuclear receptor.
- Record: found
- Abstract: found
- Article: not found