- Record: found
- Abstract: found
- Article: found
Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms
Read this article at
Abstract
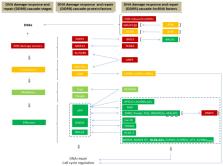
Protection of genome integrity is vital for all living organisms, particularly when DNA double-strand breaks (DSBs) occur. Eukaryotes have developed two main pathways, namely Non-Homologous End Joining (NHEJ) and Homologous Recombination (HR), to repair DSBs. While most of the current research is focused on the role of key protein players in the functional regulation of DSB repair pathways, accumulating evidence has uncovered a novel class of regulating factors termed non-coding RNAs. Non-coding RNAs have been found to hold a pivotal role in the activation of DSB repair mechanisms, thereby safeguarding genomic stability. In particular, long non-coding RNAs (lncRNAs) have begun to emerge as new players with vast therapeutic potential. This review summarizes important advances in the field of lncRNAs, including characterization of recently identified lncRNAs, and their implication in DSB repair pathways in the context of tumorigenesis.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
The transcriptional landscape of the mammalian genome.
- Record: found
- Abstract: found
- Article: not found
Gene regulation by long non-coding RNAs and its biological functions
- Record: found
- Abstract: found
- Article: not found