- Record: found
- Abstract: found
- Article: found
Transcription factor-based biosensors enlightened by the analyte
Read this article at
Abstract
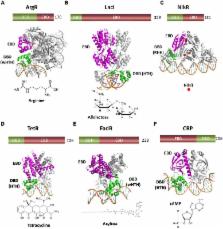
Whole cell biosensors (WCBs) have multiple applications for environmental monitoring, detecting a wide range of pollutants. WCBs depend critically on the sensitivity and specificity of the transcription factor (TF) used to detect the analyte. We describe the mechanism of regulation and the structural and biochemical properties of TF families that are used, or could be used, for the development of environmental WCBs. Focusing on the chemical nature of the analyte, we review TFs that respond to aromatic compounds (XylS-AraC, XylR-NtrC, and LysR), metal ions (MerR, ArsR, DtxR, Fur, and NikR) or antibiotics (TetR and MarR). Analyzing the structural domains involved in DNA recognition, we highlight the similitudes in the DNA binding domains (DBDs) of these TF families. Opposite to DBDs, the wide range of analytes detected by TFs results in a diversity of structures at the effector binding domain. The modular architecture of TFs opens the possibility of engineering TFs with hybrid DNA and effector specificities. Yet, the lack of a crisp correlation between structural domains and specific functions makes this a challenging task.
Related collections
Most cited references163
- Record: found
- Abstract: found
- Article: not found
Microbiological effects of sublethal levels of antibiotics.
- Record: found
- Abstract: found
- Article: not found