- Record: found
- Abstract: found
- Article: found
Genome-Wide Transcriptome Analysis Reveals Extensive Alternative Splicing Events in the Protoscoleces of Echinococcus granulosus and Echinococcus multilocularis
Read this article at
Abstract
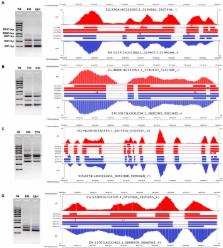
Alternative splicing (AS), as one of the most important topics in the post-genomic era, has been extensively studied in numerous organisms. However, little is known about the prevalence and characteristics of AS in Echinococcus species, which can cause significant health problems to humans and domestic animals. Based on high-throughput RNA-sequencing data, we performed a genome-wide survey of AS in two major pathogens of echinococcosis -Echinococcus granulosus and Echinococcus multilocularis. Our study revealed that the prevalence and characteristics of AS in protoscoleces of the two parasites were generally consistent with each other. A total of 6,826 AS events from 3,774 E. granulosus genes and 6,644 AS events from 3,611 E. multilocularis genes were identified in protoscolex transcriptomes, indicating that 33–36% of genes were subject to AS in the two parasites. Strikingly, intron retention instead of exon skipping was the predominant type of AS in Echinococcus species. Moreover, analysis of the Kyoto Encyclopedia of Genes and Genomes pathway indicated that genes that underwent AS events were significantly enriched in multiple pathways mainly related to metabolism (e.g., purine, fatty acid, galactose, and glycerolipid metabolism), signal transduction (e.g., Jak-STAT, VEGF, Notch, and GnRH signaling pathways), and genetic information processing (e.g., RNA transport and mRNA surveillance pathways). The landscape of AS obtained in this study will not only facilitate future investigations on transcriptome complexity and AS regulation during the life cycle of Echinococcus species, but also provide an invaluable resource for future functional and evolutionary studies of AS in platyhelminth parasites.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Next-generation transcriptome assembly.
- Record: found
- Abstract: found
- Article: not found
Alternative splicing and evolution: diversification, exon definition and function.
- Record: found
- Abstract: found
- Article: not found