- Record: found
- Abstract: found
- Article: found
The Amino-Terminal Region of Hepatitis E Virus ORF1 Containing a Methyltransferase (Met) and a Papain-Like Cysteine Protease (PCP) Domain Counteracts Type I Interferon Response
Abstract
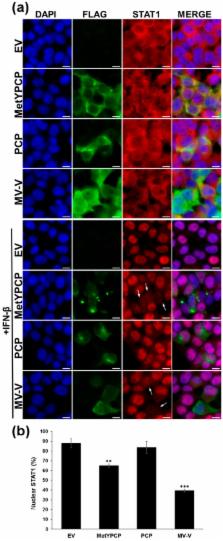
Hepatitis E virus (HEV) is responsible for large waterborne epidemics of hepatitis in endemic countries and is an emerging zoonotic pathogen worldwide. In endemic regions, HEV-1 or HEV-2 genotypes are frequently associated with fulminant hepatitis in pregnant women, while with zoonotic HEV (HEV-3 and HEV-4), chronic cases of hepatitis and severe neurological disorders are reported. Hence, it is important to characterize the interactions between HEV and its host. Here, we investigated the ability of the nonstructural polyprotein encoded by the first open reading frame ( ORF1) of HEV to modulate the host early antiviral response and, in particular, the type I interferon (IFN-I) system. We found that the amino-terminal region of HEV-3 ORF1 (MetYPCP), containing a putative methyltransferase (Met) and a papain-like cysteine protease (PCP) functional domain, inhibited IFN-stimulated response element (ISRE) promoter activation and the expression of several IFN-stimulated genes (ISGs) in response to IFN-I. We showed that the MetYPCP domain interfered with the Janus kinase (JAK)/signal transducer and activator of the transcription protein (STAT) signalling pathway by inhibiting STAT1 nuclear translocation and phosphorylation after IFN-I treatment. In contrast, MetYPCP had no effect on STAT2 phosphorylation and a limited impact on the activation of the JAK/STAT pathway after IFN-II stimulation. This inhibitory function seemed to be genotype-dependent, as MetYPCP from HEV-1 had no significant effect on the JAK/STAT pathway. Overall, this study provides evidence that the predicted MetYPCP domain of HEV ORF1 antagonises STAT1 activation to modulate the IFN response.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Interferons and viruses: an evolutionary arms race of molecular interactions.
- Record: found
- Abstract: found
- Article: not found
Viral tricks to grid-lock the type I interferon system
- Record: found
- Abstract: found
- Article: not found