- Record: found
- Abstract: found
- Article: found
Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer
Read this article at
Abstract
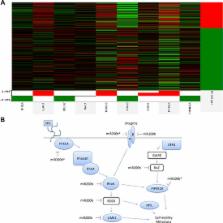
Epithelial to mesenchymal transition (EMT) involves loss of an epithelial phenotype and activation of a mesenchymal one. Enhanced expression of genes associated with a mesenchymal transition includes ZEB1/2, TWIST, and FOXC1. miRNAs are known regulators of gene expression and altered miRNA expression is known to enhance EMT in breast cancer. Here we demonstrate that the tumor suppressive miRNA family, miR-200, is not expressed in triple negative breast cancer (TNBC) cell lines and that miR-200b-3p over-expression represses EMT, which is evident through decreased migration and increased CDH1 expression. Despite the loss of migratory capacity following re-expression of miR-200b-3p, no subsequent loss of the conventional miR-200 family targets and EMT markers ZEB1/2 was observed. Next generation RNA-sequencing analysis showed that enhanced expression of pri-miR-200b lead to ectopic expression of both miR-200b-3p and miR-200b-5p with multiple isomiRs expressed for each of these miRNAs. Furthermore, miR-200b-5p was expressed in the receptor positive, epithelial breast cancer cell lines but not in the TNBC (mesenchymal) cell lines. In addition, a compensatory mechanism for miR-200b-3p/200b-5p targeting, where both miRNAs target the RHOGDI pathway leading to non-canonical repression of EMT, was demonstrated. Collectively, these data are the first to demonstrate dual targeting by miR-200b-3p and miR-200b-5p and a previously undescribed role for microRNA processing and strand expression in EMT and TNBC, the most aggressive breast cancer subtype.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Small-sample estimation of negative binomial dispersion, with applications to SAGE data.
- Record: found
- Abstract: found
- Article: not found
Moderated statistical tests for assessing differences in tag abundance.
- Record: found
- Abstract: found
- Article: not found