- Record: found
- Abstract: found
- Article: not found
Genome-wide association studies suggest sex-specific loci associated with abdominal and visceral fat
Read this article at
Abstract
Background
To identify loci associated with abdominal fat and replicate prior findings, we performed genome-wide association (GWA) studies of abdominal fat traits: subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT), total adipose tissue (TAT) and visceral to subcutaneous adipose tissue ratio (VSR).
Subjects and Methods
Sex-combined and sex-stratified analyses were performed on each trait with (TRAIT-BMI) or without (TRAIT) adjustment for BMI, and cohort-specific results were combined via a fixed effects meta-analysis. A total of 2,513 subjects of European descent were available for the discovery phase. For replication, 2,171 European Americans and 772 African Americans were available.
Results
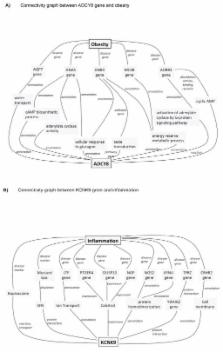
A total of 52 SNPs encompassing 7 loci showed suggestive evidence of association (p < 1.0 × 10 −6) with abdominal fat in the sex-combined analyses. The strongest evidence was found on chromosome 7p14.3 between a SNP near BBS9 gene and VAT (rs12374818; p= 1.10 × 10 −7), an association that was replicated (p = 0.02). For the BMI-adjusted trait, the strongest evidence of association was found between a SNP near CYCSP30 and VAT-BMI (rs10506943; p= 2.42 × 10 −7). Our sex-specific analyses identified one genome-wide significant (p < 5.0 × 10 −8) locus for SAT in women with 11 SNPs encompassing the MLLT10, DNAJC1 and EBLN1 genes on chromosome 10p12.31 (p = 3.97 × 10 −8 to 1.13 × 10 −8). The THNSL2 gene previously associated with VAT in women was also replicated (p= 0.006). The six gene/loci showing the strongest evidence of association with VAT or VAT-BMI were interrogated for their functional links with obesity and inflammation using the Biograph knowledge-mining software. Genes showing the closest functional links with obesity and inflammation were ADCY8 and KCNK9, respectively.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
Relation of body fat distribution to metabolic complications of obesity.
- Record: found
- Abstract: found
- Article: not found
Prioritizing GWAS results: A review of statistical methods and recommendations for their application.
- Record: found
- Abstract: found
- Article: not found
