- Record: found
- Abstract: found
- Article: found
Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing
Read this article at
Abstract
Background
Patients with prostate cancer may present with metastatic or recurrent disease despite initial curative treatment. The propensity of metastatic prostate cancer to spread to the bone has limited repeated sampling of tumor deposits. Hence, considerably less is understood about this lethal metastatic disease, as it is not commonly studied. Here we explored whole-genome sequencing of plasma DNA to scan the tumor genomes of these patients non-invasively.
Methods
We wanted to make whole-genome analysis from plasma DNA amenable to clinical routine applications and developed an approach based on a benchtop high-throughput platform, that is, Illuminas MiSeq instrument. We performed whole-genome sequencing from plasma at a shallow sequencing depth to establish a genome-wide copy number profile of the tumor at low costs within 2 days. In parallel, we sequenced a panel of 55 high-interest genes and 38 introns with frequent fusion breakpoints such as the TMPRSS2-ERG fusion with high coverage. After intensive testing of our approach with samples from 25 individuals without cancer we analyzed 13 plasma samples derived from five patients with castration resistant (CRPC) and four patients with castration sensitive prostate cancer (CSPC).
Results
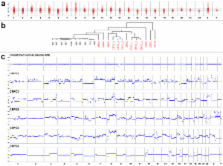
The genome-wide profiling in the plasma of our patients revealed multiple copy number aberrations including those previously reported in prostate tumors, such as losses in 8p and gains in 8q. High-level copy number gains in the AR locus were observed in patients with CRPC but not with CSPC disease. We identified the TMPRSS2-ERG rearrangement associated 3-Mbp deletion on chromosome 21 and found corresponding fusion plasma fragments in these cases. In an index case multiregional sequencing of the primary tumor identified different copy number changes in each sector, suggesting multifocal disease. Our plasma analyses of this index case, performed 13 years after resection of the primary tumor, revealed novel chromosomal rearrangements, which were stable in serial plasma analyses over a 9-month period, which is consistent with the presence of one metastatic clone.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer.
- Record: found
- Abstract: found
- Article: not found
Performance comparison of benchtop high-throughput sequencing platforms.
- Record: found
- Abstract: found
- Article: not found