- Record: found
- Abstract: found
- Article: found
A visual review of the interactome of LRRK2: Using deep-curated molecular interaction data to represent biology
Read this article at
Abstract
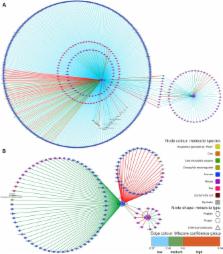
Molecular interaction databases are essential resources that enable access to a wealth of information on associations between proteins and other biomolecules. Network graphs generated from these data provide an understanding of the relationships between different proteins in the cell, and network analysis has become a widespread tool supporting –omics analysis. Meaningfully representing this information remains far from trivial and different databases strive to provide users with detailed records capturing the experimental details behind each piece of interaction evidence. A targeted curation approach is necessary to transfer published data generated by primarily low-throughput techniques into interaction databases. In this review we present an example highlighting the value of both targeted curation and the subsequent effective visualization of detailed features of manually curated interaction information. We have curated interactions involving LRRK2, a protein of largely unknown function linked to familial forms of Parkinson's disease, and hosted the data in the IntAct database. This LRRK2-specific dataset was then used to produce different visualization examples highlighting different aspects of the data: the level of confidence in the interaction based on orthogonal evidence, those interactions found under close-to-native conditions, and the enzyme–substrate relationships in different in vitro enzymatic assays. Finally, pathway annotation taken from the Reactome database was overlaid on top of interaction networks to bring biological functional context to interaction maps.
Related collections
Most cited references56

- Record: found
- Abstract: not found
- Article: not found
The cell as a collection of protein machines: preparing the next generation of molecular biologists.
- Record: found
- Abstract: found
- Article: not found