- Record: found
- Abstract: found
- Article: found
Changes in Channel Trafficking and Protein Stability Caused by LQT2 Mutations in the PAS Domain of the HERG Channel
Read this article at
Abstract
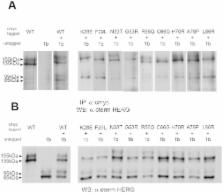
Inherited human long-QT2 syndrome (LQTS) results from mutations in the gene encoding the HERG channel. Several LQT2-associated mutations have been mapped to the amino terminal cytoplasmic Per-Arnt-Sim (PAS) domain of the HERG1a channel subunit. Here we have characterized the trafficking properties of some LQT2-associated PAS domain mutants and analyzed rescue of the trafficking mutants by low temperature (27°C) or by the pore blocker drug E4031. We show that the LQT2-associated mutations in the PAS domain of the HERG channel display molecular properties that are distinct from the properties of LQT2-associated mutations in the trans-membrane region. Unlike the latter, many of the tested PAS domain LQT2-associated mutations do not result in trafficking deficiency of the channel. Moreover, the majority of the PAS domain mutations that cause trafficking deficiencies are not rescued by a pore blocking drug. We have also explored the in vitro folding stability properties of isolated mutant PAS domain proteins using a thermal unfolding fluorescence assay and a chemical unfolding assay.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery.
- Record: found
- Abstract: found
- Article: not found
Structure and signaling mechanism of Per-ARNT-Sim domains.
- Record: found
- Abstract: found
- Article: not found