- Record: found
- Abstract: found
- Article: found
chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells
Read this article at
ABSTRACT
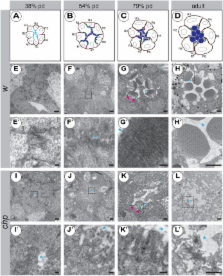
The apical surface of epithelial cells is often highly specialised to fulfil cell type-specific functions. Many epithelial cells expand their apical surface by forming microvilli, actin-based, finger-like membrane protrusions. The apical surface of Drosophila photoreceptor cells (PRCs) forms tightly packed microvilli, which are organised into the photosensitive rhabdomeres. As previously shown, the GPI-anchored adhesion protein Chaoptin is required for the stability of the microvilli, whereas the transmembrane protein Crumbs is essential for proper rhabdomere morphogenesis. Here we show that chaoptin synergises with crumbs to ensure optimal rhabdomere width. In addition, reduction of crumbs ameliorates morphogenetic defects observed in PRCs mutant for prominin and eyes shut, known antagonists of chaoptin. These results suggest that these four genes provide a balance of adhesion and anti-adhesion to maintain microvilli development and maintenance. Similar to crumbs mutant PRCs, PRCs devoid of prominin or eyes shut undergo light-dependent retinal degeneration. Given the observation that human orthologues of crumbs, prominin and eyes shut result in progressive retinal degeneration and blindness, the Drosophila eye is ideally suited to unravel the genetic and cellular mechanisms that ensure morphogenesis of PRCs and their maintenance under light-mediated stress.
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering.
- Record: found
- Abstract: found
- Article: not found
Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12).
- Record: found
- Abstract: found
- Article: not found