- Record: found
- Abstract: found
- Article: found
Speciation in the Deep Sea: Multi-Locus Analysis of Divergence and Gene Flow between Two Hybridizing Species of Hydrothermal Vent Mussels
Read this article at
Abstract
Background
Reconstructing the history of divergence and gene flow between closely-related organisms has long been a difficult task of evolutionary genetics. Recently, new approaches based on the coalescence theory have been developed to test the existence of gene flow during the process of divergence. The deep sea is a motivating place to apply these new approaches. Differentiation by adaptation can be driven by the heterogeneity of the hydrothermal environment while populations should not have been strongly perturbed by climatic oscillations, the main cause of geographic isolation at the surface.
Methodology/Principal Finding
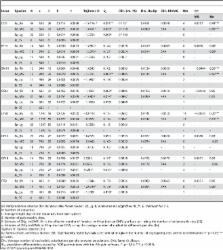
Samples of DNA sequences were obtained for seven nuclear loci and a mitochondrial locus in order to conduct a multi-locus analysis of divergence and gene flow between two closely related and hybridizing species of hydrothermal vent mussels, Bathymodiolus azoricus and B. puteoserpentis. The analysis revealed that (i) the two species have started to diverge approximately 0.760 million years ago, (ii) the B. azoricus population size was 2 to 5 time greater than the B. puteoserpentis and the ancestral population and (iii) gene flow between the two species occurred over the complete species range and was mainly asymmetric, at least for the chromosomal regions studied.
Conclusions/Significance
A long history of gene flow has been detected between the two Bathymodiolus species. However, it proved very difficult to conclusively distinguish secondary introgression from ongoing parapatric differentiation. As powerful as coalescence approaches could be, we are left by the fact that natural populations often deviates from standard assumptions of the underlying model. A more direct observation of the history of recombination at one of the seven loci studied suggests an initial period of allopatric differentiation during which recombination was blocked between lineages. Even in the deep sea, geographic isolation may well be a crucial promoter of speciation.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: found
Arlequin (version 3.0): An integrated software package for population genetics data analysis
- Record: found
- Abstract: found
- Article: not found
Dating of the human-ape splitting by a molecular clock of mitochondrial DNA.
- Record: found
- Abstract: found
- Article: not found