- Record: found
- Abstract: found
- Article: found
CC Chemokine Receptor 5: The Interface of Host Immunity and Cancer
Read this article at
Abstract
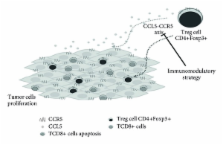
Solid tumors are embedded in a stromal microenvironment consisting of immune cells, such as macrophages and lymphocytes, as well as nonimmune cells, such as endothelial cells and fibroblasts. Chemokines are a type of small secreted chemotactic cytokine and together with their receptors play key roles in the immune defense. Critically, they regulate cancer cellular migration and also contribute to their proliferation and survival. The CCR5 chemokine receptor is involved in leucocytes chemotaxis to sites of inflammation and plays an important role in the macrophages, T cells, and monocytes recruitment. Additionally, CCR5 may have an indirect effect on cancer progression by controlling the antitumor immune response, since it has been demonstrated that its expression could promote tumor growth and contribute to tumor metastasis, in different types of malignant tumors. Furthermore, it was demonstrated that a CCR5 antagonist may inhibit tumor growth, consisting of a possible therapeutic target. In this context, the present review focuses on the establishment of CCR5 within the interface of host immunity, tumor microenvironment, and its potential as a targeting to immunotherapy.
Related collections
Most cited references89
- Record: found
- Abstract: found
- Article: not found
Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells.
- Record: found
- Abstract: found
- Article: not found
Natural regulatory T cells in infectious disease.
- Record: found
- Abstract: found
- Article: not found