- Record: found
- Abstract: found
- Article: found
Network inference with ensembles of bi-clustering trees
Read this article at
Abstract
Background
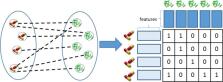
Network inference is crucial for biomedicine and systems biology. Biological entities and their associations are often modeled as interaction networks. Examples include drug protein interaction or gene regulatory networks. Studying and elucidating such networks can lead to the comprehension of complex biological processes. However, usually we have only partial knowledge of those networks and the experimental identification of all the existing associations between biological entities is very time consuming and particularly expensive. Many computational approaches have been proposed over the years for network inference, nonetheless, efficiency and accuracy are still persisting open problems. Here, we propose bi-clustering tree ensembles as a new machine learning method for network inference, extending the traditional tree-ensemble models to the global network setting. The proposed approach addresses the network inference problem as a multi-label classification task. More specifically, the nodes of a network (e.g., drugs or proteins in a drug-protein interaction network) are modelled as samples described by features (e.g., chemical structure similarities or protein sequence similarities). The labels in our setting represent the presence or absence of links connecting the nodes of the interaction network (e.g., drug-protein interactions in a drug-protein interaction network).
Results
We extended traditional tree-ensemble methods, such as extremely randomized trees (ERT) and random forests (RF) to ensembles of bi-clustering trees, integrating background information from both node sets of a heterogeneous network into the same learning framework. We performed an empirical evaluation, comparing the proposed approach to currently used tree-ensemble based approaches as well as other approaches from the literature. We demonstrated the effectiveness of our approach in different interaction prediction (network inference) settings. For evaluation purposes, we used several benchmark datasets that represent drug-protein and gene regulatory networks. We also applied our proposed method to two versions of a chemical-protein association network extracted from the STITCH database, demonstrating the potential of our model in predicting non-reported interactions.
Conclusions
Bi-clustering trees outperform existing tree-based strategies as well as machine learning methods based on other algorithms. Since our approach is based on tree-ensembles it inherits the advantages of tree-ensemble learning, such as handling of missing values, scalability and interpretability.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology

- Record: found
- Abstract: found
- Article: found
Prediction of drug–target interaction networks from the integration of chemical and genomic spaces
- Record: found
- Abstract: not found
- Article: not found