- Record: found
- Abstract: found
- Article: found
Hsp90 Inhibitors Are Efficacious against Kaposi Sarcoma by Enhancing the Degradation of the Essential Viral Gene LANA, of the Viral Co-Receptor EphA2 as well as Other Client Proteins
Read this article at
Abstract
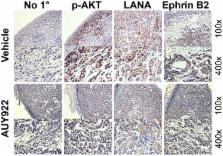
Heat-shock protein 90 (Hsp90) inhibitors exhibit activity against human cancers. We evaluated a series of new, oral bioavailable, chemically diverse Hsp90 inhibitors (PU-H71, AUY922, BIIB021, NVP-BEP800) against Kaposi sarcoma (KS). All Hsp90 inhibitors exhibited nanomolar EC 50 in culture and AUY922 reduced tumor burden in a xenograft model of KS. KS is associated with KS-associated herpesvirus (KSHV). We identified the viral latency associated nuclear antigen (LANA) as a novel client protein of Hsp90 and demonstrate that the Hsp90 inhibitors diminish the level of LANA through proteasomal degradation. These Hsp90 inhibitors also downregulated EphA2 and ephrin-B2 protein levels. LANA is essential for viral maintenance and EphA2 has recently been shown to facilitate KSHV infection; which in turn feeds latent persistence. Further, both molecules are required for KS tumor formation and both were downregulated in response to Hsp90 inhibitors. This provides a rationale for clinical testing of Hsp90 inhibitors in KSHV-associated cancers and in the eradication of latent KSHV reservoirs.
Author Summary
Heat shock proteins, such as Hsp90, aid the folding of proteins. They seem to be essential to sustain the growth of cancer cells. Hsp90 inhibitors are in clinical trials for many cancers but with mixed results, presumably since these proteins have many clients. The mechanism for drug efficacy and tumor-type variation in responses is not understood. Here we show that in the case of Kaposi sarcoma and primary effusion lymphoma, which are cancers caused by Kaposi sarcoma associated herpesvirus (KSHV/HHV8) an essential viral protein, LANA, binds to Hsp90 and is a client of Hsp90. Different small molecule Hsp90 inhibitors reduce the expression of LANA. At the same time they reduce the expression of the newly discovered co-receptor of KSHV ephA2, of Akt, cdc2 and ephrin-B2. Since LANA is required to maintain the virus latent in all tumor cells, a process, which is periodically aided by de novo infection, these inhibitors interfere with essential components of viral pathogenesis and in vivo tumor growth.
Related collections
Most cited references72
- Record: found
- Abstract: found
- Article: not found
Heat shock proteins in cancer: chaperones of tumorigenesis.
- Record: found
- Abstract: found
- Article: not found
Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone.
- Record: found
- Abstract: found
- Article: not found