- Record: found
- Abstract: found
- Article: not found
A role for the vesicle-associated tubulin binding protein ARL6 (BBS3) in flagellum extension in Trypanosoma brucei
Read this article at
Abstract
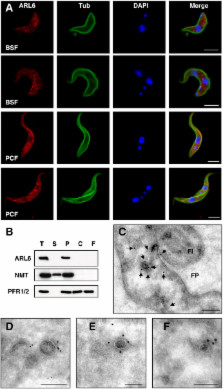
The small GTPase Arl6 is implicated in the ciliopathic human genetic disorder Bardet–Biedl syndrome, acting at primary cilia in recruitment of the octomeric BBSome complex, which is required for specific trafficking events to and from the cilium in eukaryotes. Here we describe functional characterisation of Arl6 in the flagellated model eukaryote Trypanosoma brucei, which requires motility for viability. Unlike human Arl6 which has a ciliary localisation, TbARL6 is associated with electron-dense vesicles throughout the cell body following co-translational modification by N-myristoylation. Similar to the related protein ARL-3A in T. brucei, modulation of expression of ARL6 by RNA interference does not prevent motility but causes a significant reduction in flagellum length. Tubulin is identified as an ARL6 interacting partner, suggesting that ARL6 may act as an anchor between vesicles and cytoplasmic microtubules. We provide evidence that the interaction between ARL6 and the BBSome is conserved in unicellular eukaryotes. Overexpression of BBS1 leads to translocation of endogenous ARL6 to the site of exogenous BBS1 at the flagellar pocket. Furthermore, a combination of BBS1 overexpression and ARL6 RNAi has a synergistic inhibitory effect on cell growth. Our findings indicate that ARL6 in trypanosomes contributes to flagellum biogenesis, most likely through an interaction with the BBSome.
Graphical abstract
Highlights
► The BBSome-associated protein ARL6 localises to vesicles in Trypanosoma brucei. ► T. brucei ARL6 is N-myristoylated. ► RNAi knockdown causes a decrease in flagellum length but does not affect motility. ► TbARL6 binds to tubulin and has a relatively low affinity for guanine nucleotides. ► The BBSome subunit BBS1 and ARL6 are functionally linked in trypanosomes.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei.
- Record: found
- Abstract: found
- Article: not found
Rabs and their effectors: achieving specificity in membrane traffic.

- Record: found
- Abstract: found
- Article: found

