- Record: found
- Abstract: found
- Article: found
Blood-based host biomarker diagnostics in active case finding for pulmonary tuberculosis: A diagnostic case-control study
Read this article at
Abstract
Background
There is a need to identify scalable tuberculosis screening strategies among high burden populations. The WHO has identified a non-sputum-based triage test as a development priority.
Methods
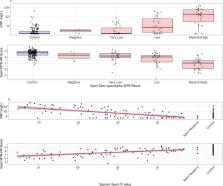
We performed a diagnostic case-control study of point-of-care C-reactive protein (CRP) and Prototype-Xpert-MTB-Host-Response (Xpert-MTB-HR) assays in the context of a mass screening program for tuberculosis in two prisons in Brazil. All incarcerated individuals irrespective of symptoms were screened by sputum Xpert MTB/RIF and sputum culture. Among consecutive, Xpert MTB/RIF or culture-confirmed cases and Xpert MTB/RIF and culture-negative controls, CRP was quantified in serum by a point-of-care assay (iChroma-II) and a 3-gene expression score was quantified from whole blood using the Xpert-MTB-HR cartridge. We evaluated receiver operating characteristic area under the curve (AUC) and assessed specificity at 90% sensitivity and sensitivity at 70% specificity, consistent with WHO target product profile (TPP) benchmarks.
Findings
Two hundred controls (no TB) and 100 culture- or Xpert MTB/RIF-positive tuberculosis cases were included. Half of tuberculosis cases and 11% of controls reported any tuberculosis symptoms. AUC for CRP was 0·79 (95% CI: 0·73–0·84) and for Xpert-MTB-HR was 0·84 (95% CI: 0·79–0·89). At 90% sensitivity, Xpert-MTB-HR had significantly higher specificity (53·0%, 95% CI: 45·0–69·0%) than CRP (28·1%, 95% CI: 20·2–41·8%) ( p = 0·003), both well below the TPP benchmark of 70%. Among individuals with medium or high sputum Xpert MTB/RIF semi-quantitative load, sensitivity (at 70% specificity) of CRP (90·3%, 95% CI: 74·2–98·0) and Xpert-MTB-HR (96·8%, 95% CI: 83·3–99·9%) was higher.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
pROC: an open-source package for R and S+ to analyze and compare ROC curves

- Record: found
- Abstract: found
- Article: found
STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies

- Record: found
- Abstract: found
- Article: found