- Record: found
- Abstract: found
- Article: not found
Binding Interactions between Receptor-Binding Domain of Spike Protein and Human Angiotensin Converting Enzyme-2 in Omicron Variant

Abstract
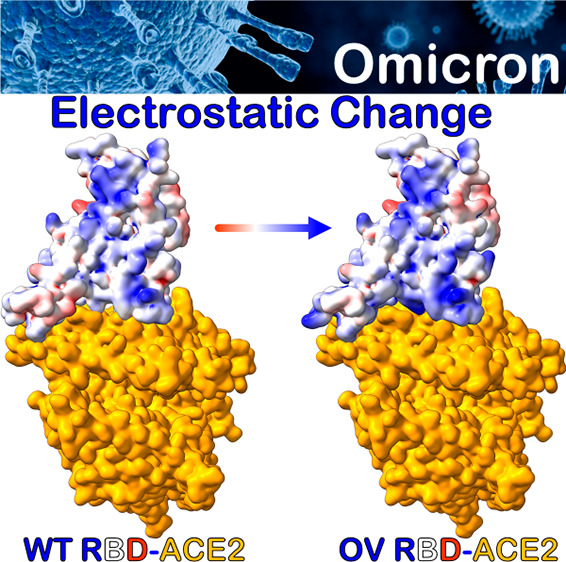
The emergence of new SARS-CoV-2 Omicron variant of concern (OV) has exacerbated the COVID-19 pandemic because of a large number of mutations in the spike protein, particularly in the receptor-binding domain (RBD), resulting in highly contagious and/or vaccine-resistant strains. Herein, we present a systematic analysis based on detailed molecular dynamics (MD) simulations in order to understand how the OV RBD mutations affect the ACE2 binding. We show that the OV RBD binds to ACE2 more efficiently and tightly predominantly because of strong electrostatic interactions, thereby promoting increased infectivity and transmissibility compared to other strains. Some of the OV RBD mutations are predicted to affect the antibody neutralization either through their role in the S-protein conformational changes, such as S371L, S373P, and S375F, or through changing its surface charge distribution, such as G339D, N440K, T478K, and E484A. Other mutations, such as K417N, G446S, and Y505H, decrease the ACE2 binding, whereas S447N, Q493R, G496S, Q498R, and N501Y tend to increase it.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: not found
- Article: not found
Comparison of simple potential functions for simulating liquid water
- Record: found
- Abstract: found
- Article: not found
