- Record: found
- Abstract: found
- Article: found
A DNA barcode library for 5,200 German flies and midges (Insecta: Diptera) and its implications for metabarcoding‐based biomonitoring

Read this article at
Abstract
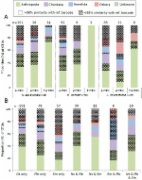
This study summarizes results of a DNA barcoding campaign on German Diptera, involving analysis of 45,040 specimens. The resultant DNA barcode library includes records for 2,453 named species comprising a total of 5,200 barcode index numbers (BINs), including 2,700 COI haplotype clusters without species‐level assignment, so called “dark taxa.” Overall, 88 out of 117 families (75%) recorded from Germany were covered, representing more than 50% of the 9,544 known species of German Diptera. Until now, most of these families, especially the most diverse, have been taxonomically inaccessible. By contrast, within a few years this study provided an intermediate taxonomic system for half of the German Dipteran fauna, which will provide a useful foundation for subsequent detailed, integrative taxonomic studies. Using DNA extracts derived from bulk collections made by Malaise traps, we further demonstrate that species delineation using BINs and operational taxonomic units (OTUs) constitutes an effective method for biodiversity studies using DNA metabarcoding. As the reference libraries continue to grow, and gaps in the species catalogue are filled, BIN lists assembled by metabarcoding will provide greater taxonomic resolution. The present study has three main goals: (a) to provide a DNA barcode library for 5,200 BINs of Diptera; (b) to demonstrate, based on the example of bulk extractions from a Malaise trap experiment, that DNA barcode clusters, labelled with globally unique identifiers (such as OTUs and/or BINs), provide a pragmatic, accurate solution to the “taxonomic impediment”; and (c) to demonstrate that interim names based on BINs and OTUs obtained through metabarcoding provide an effective method for studies on species‐rich groups that are usually neglected in biodiversity research projects because of their unresolved taxonomy.
Related collections
Most cited references113

- Record: found
- Abstract: found
- Article: not found