- Record: found
- Abstract: found
- Article: found
Production of Hybrid Chimeric PVX Particles Using a Combination of TMV and PVX-Based Expression Vectors
Read this article at
Abstract
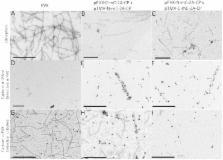
We have generated hybrid chimeric potato virus X (PVX) particles by coexpression of different PVX coat protein fusions utilizing tobacco mosaic virus (TMV) and PVX-based expression vectors. Coinfection was achieved with a modified PVX overcoat vector displaying a fluorescent protein and a TMV vector expressing another PVX fluorescent overcoat fusion protein. Coexpression of the PVX-CP fusions in the same cells was confirmed by epifluorescence microscopy. Labeling with specific antibodies and transmission electron microscopy revealed chimeric particles displaying green fluorescent protein and mCherry on the surface. These data were corroborated by bimolecular fluorescence complementation. We used split-mCherry fragments as PVX coat fusions and confirmed an interaction between the split-mCherry fragments in coinfected cells. The presence of assembled split-mCherry on the surface confirmed the hybrid character of the chimeric particles.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'.
- Record: found
- Abstract: found
- Article: not found
The 'cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring '2A-like' sequences.
- Record: found
- Abstract: not found
- Article: not found