- Record: found
- Abstract: found
- Article: not found
Multiplexed droplet single-cell RNA-sequencing using natural genetic variation
Read this article at
Abstract
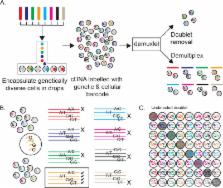
Droplet single-cell RNA-sequencing (dscRNA-seq) has enabled rapid, massively parallel profiling of transcriptomes. However, assessing differential expression across multiple individuals has been hampered by inefficient sample processing and technical batch effects. Here we describe a computational tool, demuxlet, that harnesses natural genetic variation to determine the sample identity of each cell and detect droplets containing two cells. These capabilities enable multiplexed dscRNA-seq experiments in which cells from unrelated individuals are pooled and captured at higher throughput than in standard workflows. Using simulated data, we show that 50 SNPs per cell are sufficient to assign 97% of singlets and identify 92% of doublets in pools of up to 64 individuals. Given genotyping data for each of 8 pooled samples, demuxlet correctly recovers the sample identity of >99% of singlets and identifies doublets at rates consistent with previous estimates. We apply demuxlet to assess cell type-specific changes in gene expression in 8 pooled lupus patient samples treated with IFN-β and perform eQTL analysis on 23 pooled samples.
Related collections
Most cited references21
- Record: found
- Abstract: not found
- Book: not found
Introduction to Quantitative Genetics
- Record: found
- Abstract: found
- Article: not found
Comparative Analysis of Single-Cell RNA Sequencing Methods.
- Record: found
- Abstract: found
- Article: not found
