- Record: found
- Abstract: found
- Article: found
Analysis of the developing gut microbiota in young dairy calves—impact of colostrum microbiota and gut disturbances
Read this article at
Abstract
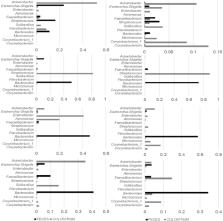
The aim of this study was to characterize the colostrum and fecal microbiota in calves and to investigate whether fecal microbiota composition was related to colostrum microbiota or factors associated with calf health. Colostrum samples were collected in buckets after hand milking of 76 calving cows from 38 smallholder dairy farms. Fecal samples were taken directly from the rectum of 76 calves at birth and at 14 days age. The bacterial community structure in colostrum and feces was analyzed by terminal restriction fragment length polymorphism for all samples, and the microbial composition was determined by 16S rRNA gene amplicon sequencing for a subset of the samples (8 colostrum, 40 fecal samples). There was a significant difference in fecal microbiota composition between day 0 and day 14 samples, but no associations between the microbiota and average daily gain, birth weight, or transfer of passive immunity. At 14 days of age, Faecalibacterium and Butyricicoccus were prevalent in higher relative abundances in the gut of healthy calves compared to calves with diarrhea that had been treated with antimicrobials. Colostrum showed great variation in composition of microbiota but no association to fecal microbiota. This study provides the first insights into the composition of colostrum and fecal microbiota of young dairy calves in southern Vietnam and can form the basis for future more detailed studies.
Related collections
Most cited references25
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found
