- Record: found
- Abstract: found
- Article: found
Knowledge Graphs of Kawasaki Disease

Read this article at
Abstract
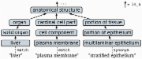
Kawasaki Disease is a vasculitis syndrome that is extremely harmful to children. Kawasaki Disease can cause severe symptoms of ischemic heart disease or develop into ischemic heart disease, leading to death in children. Researchers and clinicians need to analyze various knowledge and data resources to explore aspects of Kawasaki Disease. Knowledge Graphs have become an important AI approach to integrating various types of complex knowledge and data resources. In this paper, we present an approach for the construction of Knowledge Graphs of Kawasaki Disease. It integrates a wide range of knowledge resources related to Kawasaki Disease, including clinical guidelines, clinical trials, drug knowledge bases, medical literature, and others. It provides a basic integration foundation of knowledge and data concerning Kawasaki Disease for clinical study. In this paper, we will show that this disease-specific Knowledge Graphs are useful for exploring various aspects of Kawasaki Disease.
Related collections
Most cited references8
- Record: found
- Abstract: found
- Article: not found
An overview of MetaMap: historical perspective and recent advances.
- Record: found
- Abstract: not found
- Article: not found
From SHIQ and RDF to OWL: the making of a Web Ontology Language
- Record: found
- Abstract: found
- Article: found