- Record: found
- Abstract: found
- Article: found
Oncogene mutational profile in nasopharyngeal carcinoma
Abstract
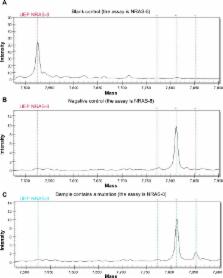
Nasopharyngeal carcinoma (NPC) is a common tumor in Southern China, but the oncogene mutational status of NPC patients has not been clarified. Using time-of-flight mass spectrometry, 238 mutation hotspots in 19 oncogenes were examined in 123 NPC patients. The relationships between mutational status and clinical data were assessed with a χ 2 or Fisher’s exact test. Survival analysis was performed using the Kaplan–Meier method with the log-rank test. In 123 patients, 21 (17.1%) NPC tumors were positive for mutations in eight oncogenes: six patients had PIK3CA mutations (4.9%), five NRAS mutations (4.1%), four KIT mutations (3.3%), two PDGFRA mutations (1.6%), two ABL mutations (1.6%), and one with simultaneous mutations in HRAS, EGFR, and BRAF (1%). Patients with mutations were more likely to relapse or develop metastasis than those with wild-type alleles ( P=0.019). No differences or correlations were found in other clinical characteristics or in patient survival. No mutations were detected in oncogenes AKT1, AKT2, CDK, ERBB2, FGFR1, FGFR3, FLT3, JAK2, KRAS, MET, and RET. These results demonstrate an association between NPC and mutations in NRAS, KIT, PIK3CA, PDGFRA, and ABL, which are associated with patient relapse and metastasis.
Most cited references42
- Record: found
- Abstract: found
- Article: not found
PDGFRA activating mutations in gastrointestinal stromal tumors.
- Record: found
- Abstract: found
- Article: not found