- Record: found
- Abstract: found
- Article: found
Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains
Read this article at
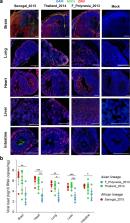
Abstract
The global emergence of Zika virus (ZIKV) revealed the unprecedented ability for a mosquito-borne virus to cause congenital birth defects. A puzzling aspect of ZIKV emergence is that all human outbreaks and birth defects to date have been exclusively associated with the Asian ZIKV lineage, despite a growing body of laboratory evidence pointing towards higher transmissibility and pathogenicity of the African ZIKV lineage. Whether this apparent paradox reflects the use of relatively old African ZIKV strains in most laboratory studies is unclear. Here, we experimentally compare seven low-passage ZIKV strains representing the recently circulating viral genetic diversity. We find that recent African ZIKV strains display higher transmissibility in mosquitoes and higher lethality in both adult and fetal mice than their Asian counterparts. We emphasize the high epidemic potential of African ZIKV strains and suggest that they could more easily go unnoticed by public health surveillance systems than Asian strains due to their propensity to cause fetal loss rather than birth defects.
Abstract
Here, the authors compare seven low passage Zika virus (ZIKV) strains representing the recently circulating viral genetic diversity of African and Asian strains and find that African ZIKV strains have higher transmissibility in mosquitoes and higher lethality in both adult and fetal mice.
Related collections
Most cited references113

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found