- Record: found
- Abstract: found
- Article: found
MicroRNA–mRNA interactions underlying colorectal cancer molecular subtypes
Read this article at
Abstract
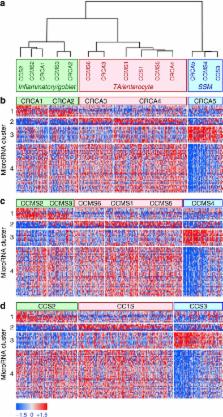
Colorectal cancer (CRC) transcriptional subtypes have been recently identified by gene expression profiling. Here we describe an analytical pipeline, microRNA master regulator analysis (MMRA), developed to search for microRNAs potentially driving CRC subtypes. Starting from a microRNA–mRNA tumour expression data set, MMRA identifies candidate regulator microRNAs by assessing their subtype-specific expression, target enrichment in subtype mRNA signatures and network analysis-based contribution to subtype gene expression. When applied to a CRC data set of 450 samples, assigned to subtypes by 3 different transcriptional classifiers, MMRA identifies 24 candidate microRNAs, in most cases downregulated in the stem/serrated/mesenchymal (SSM) poor prognosis subtype. Functional validation in CRC cell lines confirms downregulation of the SSM subtype by miR-194, miR-200b, miR-203 and miR-429, which share target genes and pathways mediating this effect. These results show that, by combining statistical tests, target prediction and network analysis, MMRA effectively identifies microRNAs functionally associated to cancer subtypes.
Abstract
 Colorectal cancer subtypes can be distinguished by their different biological and
molecular properties. Here the authors present microRNA Master Regulator Analysis,
a tool to identify microRNAs driving subtype-specific gene expression and cancer variation.
Colorectal cancer subtypes can be distinguished by their different biological and
molecular properties. Here the authors present microRNA Master Regulator Analysis,
a tool to identify microRNAs driving subtype-specific gene expression and cancer variation.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
The transcriptional network for mesenchymal transformation of brain tumors
- Record: found
- Abstract: found
- Article: not found
Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition.

- Record: found
- Abstract: found
- Article: found