- Record: found
- Abstract: found
- Article: found
An improved random forest-based computational model for predicting novel miRNA-disease associations

Read this article at
Abstract
Background
A large body of evidence shows that miRNA regulates the expression of its target genes at post-transcriptional level and the dysregulation of miRNA is related to many complex human diseases. Accurately discovering disease-related miRNAs is conductive to the exploring of the pathogenesis and treatment of diseases. However, because of the limitation of time-consuming and expensive experimental methods, predicting miRNA-disease associations by computational models has become a more economical and effective mean.
Results
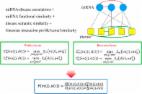
Inspired by the work of predecessors, we proposed an improved computational model based on random forest (RF) for identifying miRNA-disease associations (IRFMDA). First, the integrated similarity of diseases and the integrated similarity of miRNAs were calculated by combining the semantic similarity and Gaussian interaction profile kernel (GIPK) similarity of diseases, the functional similarity and GIPK similarity of miRNAs, respectively. Then, the integrated similarity of diseases and the integrated similarity of miRNAs were combined to represent each miRNA-disease relationship pair. Next, the miRNA-disease relationship pairs contained in the HMDD (v2.0) database were considered positive samples, and the randomly constructed miRNA-disease relationship pairs not included in HMDD (v2.0) were considered negative samples. Next, the feature selection based on the variable importance score of RF was performed to choose more useful features to represent samples to optimize the model’s ability of inferring miRNA-disease associations. Finally, a RF regression model was trained on reduced sample space to score the unknown miRNA-disease associations. The AUCs of IRFMDA under local leave-one-out cross-validation (LOOCV), global LOOCV and 5-fold cross-validation achieved 0.8728, 0.9398 and 0.9363, which were better than several excellent models for predicting miRNA-disease associations. Moreover, case studies on oesophageal cancer, lymphoma and lung cancer showed that 94 (oesophageal cancer), 98 (lymphoma) and 100 (lung cancer) of the top 100 disease-associated miRNAs predicted by IRFMDA were supported by the experimental data in the dbDEMC (v2.0) database.
Related collections
Most cited references28
- Record: found
- Abstract: not found
- Article: not found
Medical Subject Headings (MeSH).
- Record: found
- Abstract: found
- Article: not found
RWRMDA: predicting novel human microRNA-disease associations.
- Record: found
- Abstract: found
- Article: found