- Record: found
- Abstract: found
- Article: found
Probability of outbreaks and cross-border dissemination of the emerging pathogen: a genomic survey of Elizabethkingia meningoseptica
Read this article at
ABSTRACT
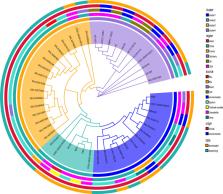
The emerging infectious agent Elizabethkingia meningoseptica is associated with life-threatening infections in immunocompromised individuals. However, there are limited data on its geographic distribution, phylogenetic evolution, pathogenesis, and transmission. In this study, we comprehensively analyze and compare the genomic features, evolutionary history, emergence date, and transmission networks of global E. meningoseptica. Geographical distribution reveals the presence of the emerging bacteria in Asia, Europe, and North America, three continents with similar latitudes. Phylogenetic analyses show no relationship between the strain’s evolutionary history and its location, origin, or source, despite the presence of genetic diversity. Analysis of the emergence timeline suggests that America is the most likely source of E. meningoseptica with the common ancestor of this pathogen dating back 90 years. Putative transmission networks indicate that E. meningoseptica bacteria can spread within the same hospital and even across borders. Minor variations in resistance genotypes and virulence genes are observed, supporting existing evidence of inherent resistance and pathogenicity in E. meningoseptica. Additionally, minocycline and doxycycline demonstrate potent antimicrobial activity against this pathogen, making them promising candidates for treating E. meningoseptica infections. Our research highlights the potential for severe nosocomial outbreaks caused by E. meningoseptica with horizontal transmission occurring between countries worldwide. To prevent future outbreak infections, increased genomic surveillance of global E. meningoseptica populations is necessary.
IMPORTANCE
Elizabethkingia meningoseptica is an emerging infectious agent associated with life-threatening infections in immunocompromised individuals. However, there are limited data available on the genomic features of E. meningoseptica. This study aims to characterize the geographical distribution, phylogenetic evolution, pathogenesis, and transmission of this bacterium. A systematic analysis of the E. meningoseptica genome revealed that a common ancestor of this bacterium existed 90 years ago. The evolutionary history showed no significant relationship with the sample source, origin, or region, despite the presence of genetic diversity. Whole genome sequencing data also demonstrated that E. meningoseptica bacteria possess inherent resistance and pathogenicity, enabling them to spread within the same hospital and even across borders. This study highlights the potential for E. meningoseptica to cause severe nosocomial outbreaks and horizontal transmission between countries worldwide. The available evidence is crucial for the development of evidence-based public health policies to prevent global outbreaks caused by emerging pathogens.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: found
RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies

- Record: found
- Abstract: found
- Article: found
High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries
- Record: found
- Abstract: found
- Article: not found