- Record: found
- Abstract: found
- Article: found
Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions
Read this article at
Abstract
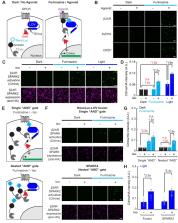
Technologies that convert transient protein-protein interactions (PPIs) into stable expression of a reporter gene are useful for genetic selections, high-throughput screening, and multiplexing with omics technologies. We previously reported SPARK (Kim et al., 2017), a transcription factor that is activated by the coincidence of blue light and a PPI. Here, we report an improved, second-generation SPARK2 that incorporates a luciferase moiety to control the light-sensitive LOV domain. SPARK2 can be temporally gated by either external light or addition of a small-molecule luciferin, which causes luciferase to open LOV via proximity-dependent BRET. Furthermore, the nested ‘AND’ gate design of SPARK2—in which both protease recruitment to the membrane-anchored transcription factor and LOV domain opening are regulated by the PPI of interest—yields a lower-background system and improved PPI specificity. We apply SPARK2 to high-throughput screening for GPCR agonists and for the detection of trans-cellular contacts, all with versatile transcriptional readout.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems.
- Record: found
- Abstract: found
- Article: not found
The genetic design of signaling cascades to record receptor activation.
- Record: found
- Abstract: found
- Article: not found