- Record: found
- Abstract: found
- Article: found
A Multi-Omics Analysis Pipeline for the Metabolic Pathway Reconstruction in the Orphan Species Quercus ilex
Read this article at
Abstract
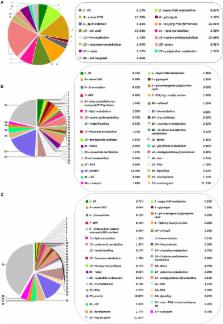
Holm oak ( Quercus ilex) is the most important and representative species of the Mediterranean forest and of the Spanish agrosilvo-pastoral “dehesa” ecosystem. Despite its environmental and economic interest, Holm oak is an orphan species whose biology is very little known, especially at the molecular level. In order to increase the knowledge on the chemical composition and metabolism of this tree species, the employment of a holistic and multi-omics approach, in the Systems Biology direction would be necessary. However, for orphan and recalcitrant plant species, specific analytical and bioinformatics tools have to be developed in order to obtain adequate quality and data-density before to coping with the study of its biology. By using a plant sample consisting of a pool generated by mixing equal amounts of homogenized tissue from acorn embryo, leaves, and roots, protocols for transcriptome (NGS-Illumina), proteome (shotgun LC-MS/MS), and metabolome (GC-MS) studies have been optimized. These analyses resulted in the identification of around 62629 transcripts, 2380 protein species, and 62 metabolites. Data are compared with those reported for model plant species, whose genome has been sequenced and is well annotated, including Arabidopsis, japonica rice, poplar, and eucalyptus. RNA and protein sequencing favored each other, increasing the number and confidence of the proteins identified and correcting erroneous RNA sequences. The integration of the large amount of data reported using bioinformatics tools allows the Holm oak metabolic network to be partially reconstructed: from the 127 metabolic pathways reported in KEGG pathway database, 123 metabolic pathways can be visualized when using the described methodology. They included: carbohydrate and energy metabolism, amino acid metabolism, lipid metabolism, nucleotide metabolism, and biosynthesis of secondary metabolites. The TCA cycle was the pathway most represented with 5 out of 10 metabolites, 6 out of 8 protein enzymes, and 8 out of 8 enzyme transcripts. On the other hand, gaps, missed pathways, included metabolism of terpenoids and polyketides and lipid metabolism. The multi-omics resource generated in this work will set the basis for ongoing and future studies, bringing the Holm oak closer to model species, to obtain a better understanding of the molecular mechanisms underlying phenotypes of interest (productive, tolerant to environmental cues, nutraceutical value) and to select elite genotypes to be used in restoration and reforestation programs, especially in a future climate change scenario.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Untargeted Metabolomics Strategies—Challenges and Emerging Directions
- Record: found
- Abstract: found
- Article: not found
Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data.
- Record: found
- Abstract: found
- Article: not found