- Record: found
- Abstract: found
- Article: found
Commensal Pseudomonas fluorescens Strains Protect Arabidopsis from Closely Related Pseudomonas Pathogens in a Colonization-Dependent Manner
Read this article at
ABSTRACT
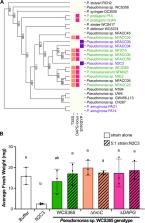
Plants form commensal associations with soil microorganisms, creating a root microbiome that provides benefits, including protection against pathogens. While bacteria can inhibit pathogens through the production of antimicrobial compounds in vitro, it is largely unknown how microbiota contribute to pathogen protection in planta. We developed a gnotobiotic model consisting of Arabidopsis thaliana and the opportunistic pathogen Pseudomonas sp. N2C3, to identify mechanisms that determine the outcome of plant-pathogen-microbiome interactions in the rhizosphere. We screened 25 phylogenetically diverse Pseudomonas strains for their ability to protect against N2C3 and found that commensal strains closely related to N2C3, including Pseudomonas sp. WCS365, were more likely to protect against pathogenesis. We used comparative genomics to identify genes unique to the protective strains and found no genes that correlate with protection, suggesting that variable regulation of components of the core Pseudomonas genome may contribute to pathogen protection. We found that commensal colonization level was highly predictive of protection, so we tested deletions in genes required for Arabidopsis rhizosphere colonization. We identified a response regulator colR, and two ColR-dependent genes with predicted roles in membrane modifications ( warB and pap2_2), that are required for Pseudomonas-mediated protection from N2C3. We found that WCS365 also protects against the agricultural pathogen Pseudomonas fuscovaginae SE-1, the causal agent of bacterial sheath brown rot of rice, in a ColR-dependent manner. This work establishes a gnotobiotic model to uncover mechanisms by which members of the microbiome can protect hosts from pathogens and informs our understanding of the use of beneficial strains for microbiome engineering in dysbiotic soil systems.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
The global burden of pathogens and pests on major food crops
- Record: found
- Abstract: found
- Article: not found
Structure and functions of the bacterial microbiota of plants.
- Record: found
- Abstract: found
- Article: not found