- Record: found
- Abstract: found
- Article: not found
Phaser crystallographic software
Read this article at
Abstract
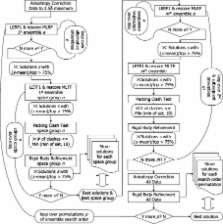
A description is given of Phaser-2.1: software for phasing macromolecular crystal structures by molecular replacement and single-wavelength anomalous dispersion phasing.
Abstract
Phaser is a program for phasing macromolecular crystal structures by both molecular replacement and experimental phasing methods. The novel phasing algorithms implemented in Phaser have been developed using maximum likelihood and multivariate statistics. For molecular replacement, the new algorithms have proved to be significantly better than traditional methods in discriminating correct solutions from noise, and for single-wavelength anomalous dispersion experimental phasing, the new algorithms, which account for correlations between F + and F −, give better phases (lower mean phase error with respect to the phases given by the refined structure) than those that use mean F and anomalous differences Δ F. One of the design concepts of Phaser was that it be capable of a high degree of automation. To this end, Phaser (written in C++) can be called directly from Python, although it can also be called using traditional CCP4 keyword-style input. Phaser is a platform for future development of improved phasing methods and their release, including source code, to the crystallographic community.
