- Record: found
- Abstract: found
- Article: found
Bacterial Species Associated With Human Inflammatory Bowel Disease and Their Pathogenic Mechanisms
Read this article at
Abstract
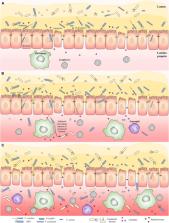
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal tract with unknown etiology. The pathogenesis of IBD results from immune responses to microbes in the gastrointestinal tract. Various bacterial species that are associated with human IBD have been identified. However, the microbes that trigger the development of human IBD are still not clear. Here we review bacterial species that are associated with human IBD and their pathogenic mechanisms to provide an updated broad understanding of this research field. IBD is an inflammatory syndrome rather than a single disease. We propose a three-stage pathogenesis model to illustrate the roles of different IBD-associated bacterial species and gut commensal bacteria in the development of human IBD. Finally, we recommend microbe-targeted therapeutic strategies based on the three-stage pathogenesis model.
Related collections
Most cited references120
- Record: found
- Abstract: found
- Article: not found
Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies.

- Record: found
- Abstract: found
- Article: found
Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases
- Record: found
- Abstract: found
- Article: found