- Record: found
- Abstract: found
- Article: found
Visceral Adipose Tissue Phospholipid Signature of Insulin Sensitivity and Obesity

Read this article at
Abstract
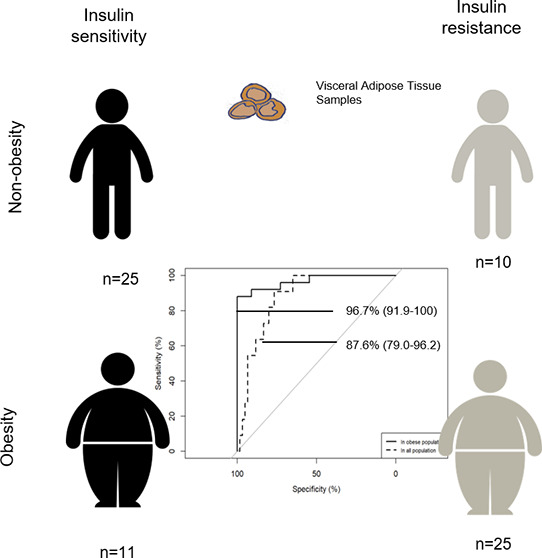
Alterations in visceral adipose tissue (VAT) are closely linked to cardiometabolic abnormalities. The aim of this work is to define a metabolic signature in VAT of insulin resistance (IR) dependent on, and independent of, obesity. An untargeted UPLC-Q-Exactive metabolomic approach was carried out on the VAT of obese insulin-sensitive (IS) and insulin-resistant subjects ( N = 11 and N = 25, respectively) and nonobese IS and IR subjects ( N = 25 and N = 10, respectively). The VAT metabolome in obesity was defined among other things by changes in the metabolism of lipids, nucleotides, carbohydrates, and amino acids, whereas when combined with high IR, it affected the metabolism of 18 carbon fatty acyl-containing phospholipid species. A multimetabolite model created by glycerophosphatidylinositol (18:0); glycerophosphatidylethanolamine (18:2); glycerophosphatidylserine (18:0); and glycerophosphatidylcholine (18:0/18:1), (18:2/18:2), and (18:2/18:3) exhibited a highly predictive performance to identify the metabotype of “insulin-sensitive obesity” among obese individuals [area under the curve (AUC) 96.7% (91.9–100)] and within the entire study population [AUC 87.6% (79.0–96.2)]. We demonstrated that IR has a unique and shared metabolic signature dependent on, and independent of, obesity. For it to be used in clinical practice, these findings need to be validated in a more accessible sample, such as blood.
Related collections
Most cited references29
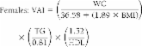
- Record: found
- Abstract: found
- Article: found
Visceral Adiposity Index

- Record: found
- Abstract: found
- Article: found
Organization of GC/MS and LC/MS metabolomics data into chemical libraries
- Record: found
- Abstract: found
- Article: not found