- Record: found
- Abstract: found
- Article: found
The role of histone methyltransferases in neurocognitive disorders associated with brain size abnormalities
Read this article at
Abstract
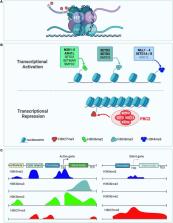
Brain size is controlled by several factors during neuronal development, including neural progenitor proliferation, neuronal arborization, gliogenesis, cell death, and synaptogenesis. Multiple neurodevelopmental disorders have co-morbid brain size abnormalities, such as microcephaly and macrocephaly. Mutations in histone methyltransferases that modify histone H3 on Lysine 36 and Lysine 4 (H3K36 and H3K4) have been identified in neurodevelopmental disorders involving both microcephaly and macrocephaly. H3K36 and H3K4 methylation are both associated with transcriptional activation and are proposed to sterically hinder the repressive activity of the Polycomb Repressor Complex 2 (PRC2). During neuronal development, tri-methylation of H3K27 (H3K27me3) by PRC2 leads to genome wide transcriptional repression of genes that regulate cell fate transitions and neuronal arborization. Here we provide a review of neurodevelopmental processes and disorders associated with H3K36 and H3K4 histone methyltransferases, with emphasis on processes that contribute to brain size abnormalities. Additionally, we discuss how the counteracting activities of H3K36 and H3K4 modifying enzymes vs. PRC2 could contribute to brain size abnormalities which is an underexplored mechanism in relation to brain size control.
Related collections
Most cited references271
- Record: found
- Abstract: found
- Article: not found
Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism
- Record: found
- Abstract: found
- Article: not found
High-resolution profiling of histone methylations in the human genome.
- Record: found
- Abstract: found
- Article: not found