- Record: found
- Abstract: found
- Article: found
Programming multicellular assembly with synthetic cell adhesion molecules
Read this article at
Abstract
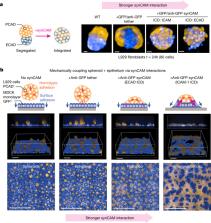
Cell adhesion molecules are ubiquitous in multicellular organisms, specifying precise cell–cell interactions in processes as diverse as tissue development, immune cell trafficking and the wiring of the nervous system 1– 4 . Here we show that a wide array of synthetic cell adhesion molecules can be generated by combining orthogonal extracellular interactions with intracellular domains from native adhesion molecules, such as cadherins and integrins. The resulting molecules yield customized cell–cell interactions with adhesion properties that are similar to native interactions. The identity of the intracellular domain of the synthetic cell adhesion molecules specifies interface morphology and mechanics, whereas diverse homotypic or heterotypic extracellular interaction domains independently specify the connectivity between cells. This toolkit of orthogonal adhesion molecules enables the rationally programmed assembly of multicellular architectures, as well as systematic remodelling of native tissues. The modularity of synthetic cell adhesion molecules provides fundamental insights into how distinct classes of cell–cell interfaces may have evolved. Overall, these tools offer powerful abilities for cell and tissue engineering and for systematically studying multicellular organization.
Abstract
Synthetic cell adhesion molecules yield customized cell–cell interactions with adhesion properties that are similar to native interactions, and offer abilities for cell and tissue engineering and for systematically studying multicellular organization.
Related collections
Most cited references63

- Record: found
- Abstract: found
- Article: found
UniProt: the universal protein knowledgebase in 2021
- Record: found
- Abstract: found
- Article: not found
Chimeric antigen receptor-modified T cells for acute lymphoid leukemia.
- Record: found
- Abstract: found
- Article: not found