- Record: found
- Abstract: found
- Article: not found
c-Jun N-terminal kinase–mediated Rad18 phosphorylation facilitates Polη recruitment to stalled replication forks
Read this article at
Abstract
The association of Rad18 with Polη is crucial for efficient translesion synthesis and DNA damage tolerance. Rad18–Polη interactions and UV tolerance depend on JNK-dependent Rad18 phosphorylation. These results provide a new mechanism by which SAPK signaling promotes genome maintenance.
Abstract
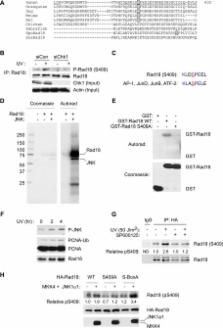
The E3 ubiquitin ligase Rad18 chaperones DNA polymerase η (Polη) to sites of UV-induced DNA damage and monoubiquitinates proliferating cell nuclear antigen (PCNA), facilitating engagement of Polη with stalled replication forks and promoting translesion synthesis (TLS). It is unclear how Rad18 activities are coordinated with other elements of the DNA damage response. We show here that Ser-409 residing in the Polη-binding motif of Rad18 is phosphorylated in a checkpoint kinase 1–dependent manner in genotoxin-treated cells. Recombinant Rad18 was phosphorylated specifically at S409 by c-Jun N-terminal kinase (JNK) in vitro. In UV-treated cells, Rad18 S409 phosphorylation was inhibited by a pharmacological JNK inhibitor. Conversely, ectopic expression of JNK and its upstream kinase mitogen-activated protein kinase kinase 4 led to DNA damage–independent Rad18 S409 phosphorylation. These results identify Rad18 as a novel JNK substrate. A Rad18 mutant harboring a Ser → Ala substitution at S409 was compromised for Polη association and did not redistribute Polη to nuclear foci or promote Polη−PCNA interaction efficiently relative to wild-type Rad18. Rad18 S409A also failed to fully complement the UV sensitivity of Rad18-depleted cells. Taken together, these results show that Rad18 phosphorylation by JNK represents a novel mechanism for promoting TLS and DNA damage tolerance.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis.
- Record: found
- Abstract: found
- Article: not found
Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function.
- Record: found
- Abstract: found
- Article: not found
