- Record: found
- Abstract: found
- Article: found
Preservation of memory B cell homeostasis in an individual producing broadly neutralising antibodies against HIV-1
Read this article at
Abstract
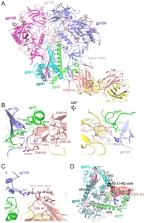
Immunological determinants favouring emergence of broadly neutralising antibodies are crucial to the development of HIV-1 vaccination strategies. Here, we combined RNAseq and B cell cloning approaches to isolate a broadly neutralising antibody (bnAb) ELC07 from an individual living with untreated HIV-1. Using single particle cryogenic electron microscopy (cryo-EM), we show that the antibody recognises a conformational epitope at the gp120-gp41 interface. ELC07 binds the closed state of the viral glycoprotein causing considerable perturbations to the gp41 trimer core structure. Phenotypic analysis of memory B cell subsets from the ELC07 bnAb donor revealed a lack of expected HIV-1-associated dysfunction, specifically no increase in CD21 −/CD27 − cells was observed whilst the resting memory (CD21 +/CD27 +) population appeared preserved despite uncontrolled HIV-1 viraemia. Moreover, single cell transcriptomes of memory B cells from this bnAb donor showed a resting memory phenotype irrespective of the epitope they targeted or their ability to neutralise diverse strains of HIV-1. Strikingly, single memory B cells from the ELC07 bnAb donor were transcriptionally similar to memory B cells from HIV-negative individuals. Our results demonstrate that potent bnAbs can arise without the HIV-1-induced dysregulation of the memory B cell compartment and suggest that sufficient levels of antigenic stimulation with a strategically designed immunogen could be effective in HIV-negative vaccine recipients.
Related collections
Most cited references116
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.

- Record: found
- Abstract: found
- Article: found
RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome
- Record: found
- Abstract: found
- Article: not found