- Record: found
- Abstract: found
- Article: found
Proteomic evidence of dietary sources in ancient dental calculus
Read this article at
Abstract
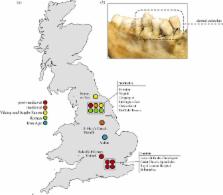
Archaeological dental calculus has emerged as a rich source of ancient biomolecules, including proteins. Previous analyses of proteins extracted from ancient dental calculus revealed the presence of the dietary milk protein β-lactoglobulin, providing direct evidence of dairy consumption in the archaeological record. However, the potential for calculus to preserve other food-related proteins has not yet been systematically explored. Here we analyse shotgun metaproteomic data from 100 archaeological dental calculus samples ranging from the Iron Age to the post-medieval period (eighth century BC to nineteenth century AD) in England, as well as 14 dental calculus samples from contemporary dental patients and recently deceased individuals, to characterize the range and extent of dietary proteins preserved in dental calculus. In addition to milk proteins, we detect proteomic evidence of foodstuffs such as cereals and plant products, as well as the digestive enzyme salivary amylase. We discuss the importance of optimized protein extraction methods, data analysis approaches and authentication strategies in the identification of dietary proteins from archaeological dental calculus. This study demonstrates that proteomic approaches can robustly identify foodstuffs in the archaeological record that are typically under-represented due to their poor macroscopic preservation.
Related collections
Most cited references47

- Record: found
- Abstract: found
- Article: found
The Proteomics Identifications (PRIDE) database and associated tools: status in 2013
- Record: found
- Abstract: found
- Article: not found
Diet and the evolution of human amylase gene copy number variation.
- Record: found
- Abstract: found
- Article: not found