- Record: found
- Abstract: found
- Article: found
Artificial Selection of Gn1a Plays an Important role in Improving Rice Yields Across Different Ecological Regions
Read this article at
Abstract
Background
Rice is one of the most important crops, and it is essential to improve rice productivity to satisfy the future global food supply demands. Gn1a ( OsCKX2), which encodes cytokinin oxidase/dehydrogenase, plays an important role in regulating rice grain yield.
Results
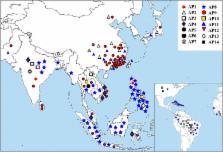
In this study, we analyzed the genetic variation of Gn1a, which influences grain yield through controlling the number of spikelets in rice. The allelic variations in the promoter, 5’ untranslated region (UTR) and coding sequence (CDS) of Gn1a were investigated in 175 cultivars and 21 wild rice accessions. We found that Gn1a showed less sequence variation in the cultivars, but exhibited significant nucleotide diversity in wild rice. A total of 14 alleles, named AP1 to AP14, were identified in the cultivars based on the amino acid divergence of GN1A. Association analysis revealed that the number of spikelets and grain yield were significantly different between the different alleles. Phylogenetic analysis indicated that the three main alleles, AP3, AP8 and AP9, in the cultivars might originate from a common ancestor allele, AP1, in wild rice.
Conclusions
Of these alleles in the cultivars, AP9 was suggested as the best allele in indica, as it has shown strong artificial selection in breeding high-yield rice in the past. It might be valuable to explore the high-yield-related alleles of Gn1a to develop high-yield rice cultivars in future breeding programs.
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
Cytokinin oxidase regulates rice grain production.
- Record: found
- Abstract: found
- Article: not found
Natural variation at the DEP1 locus enhances grain yield in rice.
- Record: found
- Abstract: found
- Article: not found