- Record: found
- Abstract: found
- Article: found
Development of nutrigenomic based precision management model for Hanwoo steers
Read this article at
Abstract
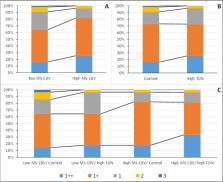
Focusing high marble deposition, Hanwoo feedlot system uses high-energy diet over the prolonged fattening period. However, due to the individual genetic variation, around 40% of them are graded into inferior quality grades (QG), despite they utilized the same resources. Therefore, focusing on development of a nutrigenomic based precision management model, this study was to evaluated the response to the divergent selection on genetic merit for marbling score (MS), under different dietary total digestible nutrient (TDN) levels. Total of 111 calves were genotyped and initially grouped according to estimated breeding value (high and low) for marbling score (MS-EBV). Subsequently, managed under two levels of feed TDN%, over the calf period, early, middle, and final fattening periods following 2 × 2 factorial arrangement. Carcasses were evaluated for MS, Back fat thickness (BFT) and Korean beef quality grading standard. As the direct response to the selection was significant, the results confirmed the importance of initial genetic grouping of Hanwoo steers for MS-EBV. However, dietary TDN level did not show an effect ( p > 0.05) on the MS. Furthermore, no genetic-by-nutrition interaction for MS ( p > 0.05) was also observed. The present results showed no correlation response on BFT ( p > 0.05), which indicates that the selection based on MS-EBV can be used to enhance the MS without undesirable effect on BFT. Ultimate turnover of the Hanwoo feedlot operation is primarily determined by the QGs. The present model shows that the initial grouping for MS-EBV increased the proportion of carcasses graded for higher QGs (QG1 ++ and QG1 +) by approximately 20%. Moreover, there appear to be a potential to increase the proportion of QG 1 ++ animals among the high-genetic group by further increasing the dietary energy content. Overall, this precision management strategy suggests the importance of adopting an MS based initial genetic grouping system for Hanwoo steers with a subsequent divergent management based on dietary energy level.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
PLINK: a tool set for whole-genome association and population-based linkage analyses.
- Record: found
- Abstract: found
- Article: not found
GCTA: a tool for genome-wide complex trait analysis.
- Record: found
- Abstract: found
- Article: not found