- Record: found
- Abstract: found
- Article: not found
Superspreading and the effect of individual variation on disease emergence
Read this article at
Coughs and sneezes...
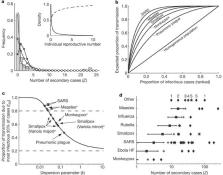
From Typhoid Mary to SARS, it has long been known that some people spread disease more than others. But for diseases transmitted via casual contact, contagiousness arises from a plethora of social and physiological factors, so epidemiologists have tended to rely on population averages to assess a disease's potential to spread. A new analysis of outbreak data shows that individual differences in infectiousness exert powerful influences on the epidemiology of ten deadly diseases. SARS and measles (and perhaps avian influenza) show strong tendencies towards ‘superspreading events’ that can ignite explosive epidemics — but this same volatility makes outbreaks more likely to fizzle out. Smallpox and pneumonic plague, two potential bioterrorism agents, show steadier growth but still differ markedly from the traditional average-based view. These findings are relevant to how emerging diseases are detected and controlled.
Abstract
Population-level analyses often use average quantities to describe heterogeneous systems, particularly when variation does not arise from identifiable groups 1, 2 . A prominent example, central to our current understanding of epidemic spread, is the basic reproductive number, R 0, which is defined as the mean number of infections caused by an infected individual in a susceptible population 3, 4 . Population estimates of R 0 can obscure considerable individual variation in infectiousness, as highlighted during the global emergence of severe acute respiratory syndrome (SARS) by numerous ‘superspreading events’ in which certain individuals infected unusually large numbers of secondary cases 5, 6, 7, 8, 9, 10 . For diseases transmitted by non-sexual direct contacts, such as SARS or smallpox, individual variation is difficult to measure empirically, and thus its importance for outbreak dynamics has been unclear 2, 10, 11 . Here we present an integrated theoretical and statistical analysis of the influence of individual variation in infectiousness on disease emergence. Using contact tracing data from eight directly transmitted diseases, we show that the distribution of individual infectiousness around R 0 is often highly skewed. Model predictions accounting for this variation differ sharply from average-based approaches, with disease extinction more likely and outbreaks rarer but more explosive. Using these models, we explore implications for outbreak control, showing that individual-specific control measures outperform population-wide measures. Moreover, the dramatic improvements achieved through targeted control policies emphasize the need to identify predictive correlates of higher infectiousness. Our findings indicate that superspreading is a normal feature of disease spread, and to frame ongoing discussion we propose a rigorous definition for superspreading events and a method to predict their frequency.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Network theory and SARS: predicting outbreak diversity
- Record: found
- Abstract: not found
- Article: not found
Heterogeneities in the transmission of infectious agents: Implications for the design of control programs
- Record: found
- Abstract: found
- Article: not found
