- Record: found
- Abstract: found
- Article: found
Rheumatoid Arthritis and miRNAs: A Critical Review through a Functional View
Read this article at
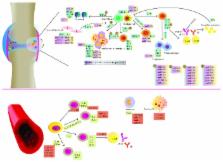
Abstract
Rheumatoid arthritis (RA) is a systemic autoimmune disease with severe joint inflammation and destruction associated with an inflammatory environment. The etiology behind RA remains to be elucidated; most updated concepts include the participation of environmental, proteomic, epigenetic, and genetic factors. Epigenetic is considered the missing link to explain genetic diversification among RA patients. Within epigenetic factors participating in RA, miRNAs are defined as small noncoding molecules with a length of approximately 22 nucleotides, capable of gene expression modulation, either negatively through inhibition of translation and degradation of the mRNA or positively through increasing the translation rate. Over the last decade and due to the feasibility of the identification of miRNAs among different tissues and compartments, they have been proposed as biomarkers for diagnosis, prognosis, and response to treatment in different pathologies. Nevertheless, miRNAs seem to be important regulators of networks instead of single genes; their hypothetical use as biomarkers needs to rely on a functional integrative description of their effects in the biological process of autoimmune conditions which until now is missing. Therefore, we underwent a bibliographic search for review and original articles related to miRNAs and their possible implications in rheumatoid arthritis. We found 48 different studies using the key words “miRNAs” or “micro-RNAs” and “rheumatoid arthritis” with restriction of publication dates from 2011 to 2016, in humans, using the English language. After a critical reading, we provide in this paper a functional view with respect to miRNA biogenesis, interaction with targets that are expressed in specific cells and tissues, during different stages of inflammatory responses associated with RA, and recognized specific areas where miRNAs might also have a pathogenic role but remain undescribed. Our results will be useful in designing future research projects that can support miRNAs as biomarkers or therapeutic targets in RA.
Related collections
Most cited references70

- Record: found
- Abstract: found
- Article: found
An Integrated Encyclopedia of DNA Elements in the Human Genome
- Record: found
- Abstract: found
- Article: not found