- Record: found
- Abstract: found
- Article: found
Identification of hepatic fibrosis inhibitors through morphometry analysis of a hepatic multicellular spheroids model
Read this article at
Abstract
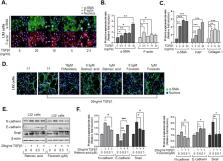
A chronic, local inflammatory milieu can cause tissue fibrosis that results in epithelial-to-mesenchymal transition (EMT), endothelial-to-mesenchymal transition (EndMT), increased abundance of fibroblasts, and further acceleration of fibrosis. In this study, we aimed to identify potential mechanisms and inhibitors of fibrosis using 3D model-based phenotypic screening. We established liver fibrosis models using multicellular tumor spheroids (MCTSs) composed of hepatocellular carcinoma (HCC) and stromal cells such as fibroblasts (WI38), hepatic stellate cells (LX2), and endothelial cells (HUVEC) seeded at constant ratios. Through high-throughput screening of FDA-approved drugs, we identified retinoic acid and forskolin as candidates to attenuate the compactness of MCTSs as well as inhibit the expression of ECM-related proteins. Additionally, retinoic acid and forskolin induced reprogramming of fibroblast and cancer stem cells in the HCC microenvironment. Of interest, retinoic acid and forskolin had anti-fibrosis effects by decreasing expression of α-SMA and F-actin in LX2 cells and HUVEC cells. Moreover, when sorafenib was added along with retinoic acid and forskolin, apoptosis was increased, suggesting that anti-fibrosis drugs may improve tissue penetration to support the efficacy of anti-cancer drugs. Collectively, these findings support the potential utility of morphometric analyses of hepatic multicellular spheroid models in the development of new drugs with novel mechanisms for the treatment of hepatic fibrosis and HCCs.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Liver fibrosis.

- Record: found
- Abstract: found
- Article: found
Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning
- Record: found
- Abstract: found
- Article: not found