- Record: found
- Abstract: found
- Article: found
Microbiome Big-Data Mining and Applications Using Single-Cell Technologies and Metagenomics Approaches Toward Precision Medicine
Read this article at
Abstract
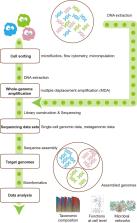
With the development of high-throughput sequencing technologies as well as various bioinformatics analytic tools, microbiome is not a “microbial dark matter” anymore. In this review, we first summarized the current analytical strategies used for big-data mining such as single-cell sequencing and metagenomics. We then provided insights into the integration of these strategies, showing significant advantages in fully describing microbiome from multiple aspects. Moreover, we discussed the correlation between gut microbiome with host organs and diseases, confirming the importance of big-data mining in clinical practices. We finally proposed new ideas about the trend of big-data mining in microbiome using multi-omics approaches and single-cell sequencing. The integration of multi-omics approaches and single-cell sequencing can provide full understanding of microbiome at both macroscopic level and microscopic level, thus contributing to precision medicine.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease.
- Record: found
- Abstract: found
- Article: not found
Commensal host-bacterial relationships in the gut.
- Record: found
- Abstract: found
- Article: not found