- Record: found
- Abstract: found
- Article: found
Identification of Philaster apodigitiformis in aquaculture and functional characterization of its β-PKA gene and expression analysis of infected Poecilia reticulata
Read this article at
Abstract
Abstract
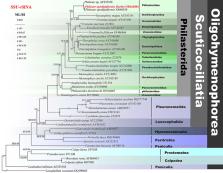
Cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA) is a distinctive member of the serine–threonine protein AGC kinase family and an effective kinase for cAMP signal transduction. In recent years, scuticociliate has caused a lot of losses in domestic fishery farming, therefore, we have carried out morphological and molecular biological studies. In this study, diseased guppies ( Poecilia reticulata) were collected from an ornamental fish market, and scuticociliate Philaster apodigitiformis Miao et al., 2009 was isolated. In our prior transcriptome sequencing research, we discovered significant expression of the β-PKA gene in P. apodigitiformis during its infection process, leading us to speculate its involvement in pathogenesis. A complete sequence of the β-PKA gene was cloned, and quantified by quantitative reverse transcription-polymerase chain reaction to analyse or to evaluate the functional characteristics of the β-PKA gene. Morphological identification and phylogenetic analysis based on small subunit rRNA sequence, infection experiments and haematoxylin–eosin staining method were also carried out, in order to study the pathological characteristics and infection mechanism of scuticociliate. The present results showed that: (1) our results revealed that β-PKA is a crucial gene involved in P. apodigitiformis infection in guppies, and the findings provide valuable insights for future studies on scuticociliatosis; (2) we characterized a complete gene, β-PKA, that is generally expressed in parasitic organisms during infection stage and (3) the present study indicates that PKA plays a critical role in scuticociliate when infection occurs by controlling essential steps such as cell growth, development and regulating the activity of the sensory body structures and the irritability system.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
MrBayes 3: Bayesian phylogenetic inference under mixed models.
- Record: found
- Abstract: found
- Article: not found
A rapid bootstrap algorithm for the RAxML Web servers.
- Record: found
- Abstract: not found
- Conference Proceedings: not found
